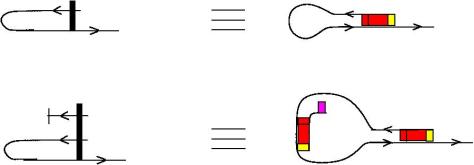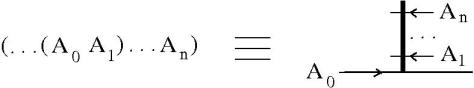UPDATE: This post is superseded by a newer one. The transcript is available here, in the chorasimilarity telegram channel (but you don’t need telegram to see it). In case you do use telegram, here is the chorasimilarity channel of long reads.
Also available on github.
In article form is on figshare.
And finally here as pdf.
___
This is the transcript of the talk from September 2020, which I gave at the Quantum Topology Seminar organized by Louis Kauffman.
There is a video of the talk and a github repository with photos of slides.
My problem is if we can compute with tangles and the R moves. I am going to tell you where does this problem comes from my point of view, why it is different than the usual way of using tangles in computation and then I’m going to propose you an improvement of the thing called zipper logic, namely the way to do universal computation using tangles and zippers.
The problem is to COMPUTE with Reidemeister moves. The problem is to use a graph rewrite system which contains the tangle diagrams and as graph rewrites the R moves.
Can we do universal computation with this?
Where does this problem come from?
This is not the usual way to do computation with tangles. The usual way is that we have a circuit which we represent as a tangle, a knot diagram, where the crossings and maybe caps and cups are operators which satisfy the algebraic equivalent of the R moves. The circuit represents a computation. When we have two circuits, then we can say that they represent equivalent computations when we can turn one into another by using R moves.
In a quantum computation we have preparation (cups) and detectors (caps) (Louis).
R moves transform a computation into another. Example with teleportation.
R moves don’t compute, they are used to prove that 2 computations are equivalent. This is not what I’m after.
The source of interest: emergent algebras. An e.a. is a combination of algebraic and topological information…
[See https://mbuliga.github.io/colin/colin.pdf as a better source for a short description of emergent algebras]
We can represent graphically the axioms. This is the configuration which gives the approximate sum. It says that as epsilon goes to 0 you obtain in the limit some gate which has 3 inputs and 3 outputs.
We say that the sum is an emergent object, it emerges from the limit. We can also define in the same way differentiability.
We can define not only emergent objects, but also emergent properties.
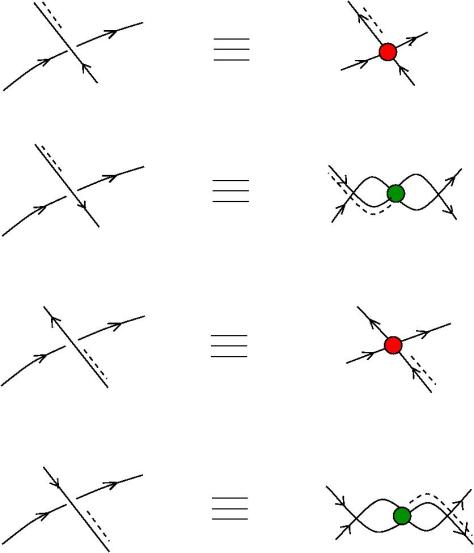
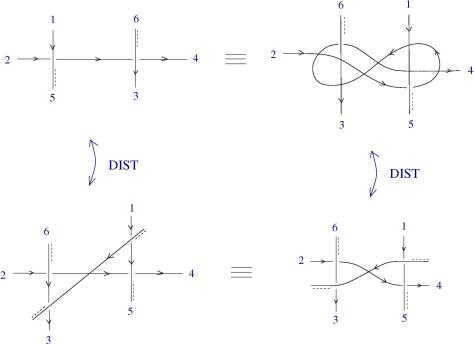
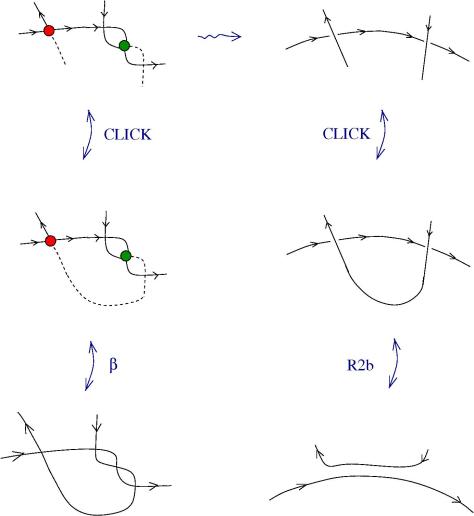
Here you see an example: we use the R2 rewrites and passage to the limit to prove that the addition is commutative.
The moral is: you pass to the limit here and there, then this becomes by a passage to the limit the associativity of the operation.
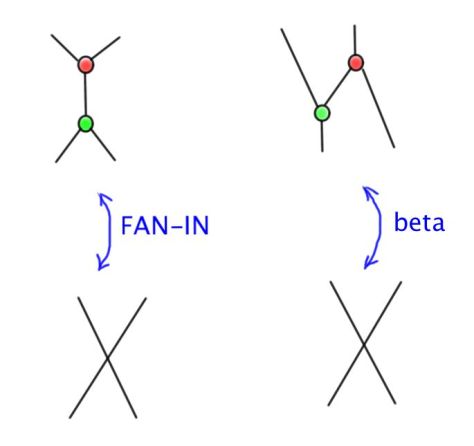
Another example: if you define a new crossing (relative crossing) then you can pass to the limit and you can prove that, based on e.a. axioms. Moreover you can prove, by using only R1 and R2 and passage to the limit, that the R3 emerges from R1, R2 and a passage to the limit, for the relative crossings.
With e.a. we can do differential calculus. We use only R1, R2 and a passage to the limit. It is a differential calculus which is not commutative.
There are interesting classes of e.a.:
- linear e.a. correspond to calculus on conical groups (Carnot groups, explanations)
- commutative e.a. which satisfy SHUFFLE (calculus in topological vector spaces) In this class you can do any computation (Pure See https://mbuliga.github.io/quinegraphs/puresee.html )
What I want to know is: can you do universal computation in the linear case? This corresponds to the initial problem.
What means universal computation? There are many, but among them, 3 ways to define what computation means. They are equivalent in a precise sense.
Lambda calculus is a term rewrite system (follows definition of lambda calculus).
Turing machine is an automaton (follows definition of TM).
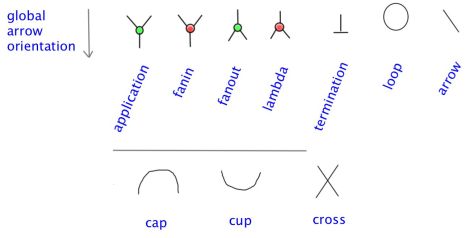
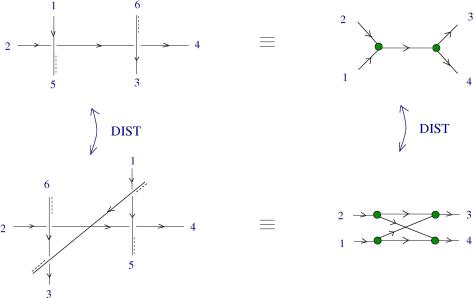
Lafont’ Interaction combinators is a graph rewrite system, where you use graphs with two types of 3valent nodes and one type of 1valent node. Explanation of rewrites. These are port graphs.
Lafont proves that the grs is universal because he can implement TM, so it has to be universal. There is a lot of work to implement LC in a grs, but the reality is that this is extremely dificult, in the sense that there are solutions, but the solutions are not what we want, in the sense that you can transform a lambda term into a graph and then reduce it with the grs of Lafont, say, and then you can decorate the edges of the graph so that you can retrieve the result of the computation. But these are non-local. (Explanation of local)
We have 3 definition of what computation means, by 3 different models, which are equivalent only if you add supplementary hypotheses. For me IC is the most important one, but we don’t know yet how to compute with IC.
Let me reformulate the problem of if we can compute with R moves in this way.
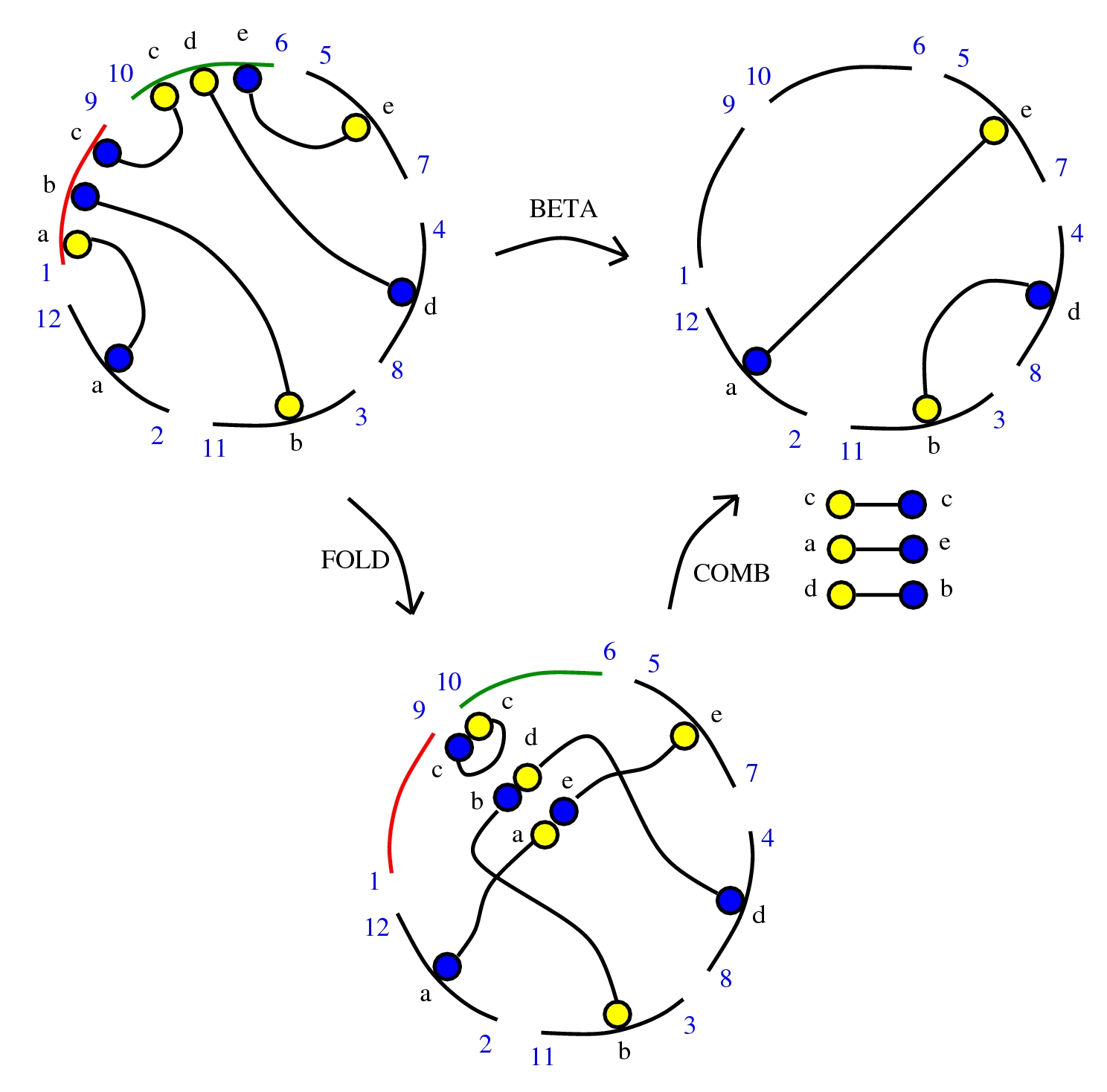
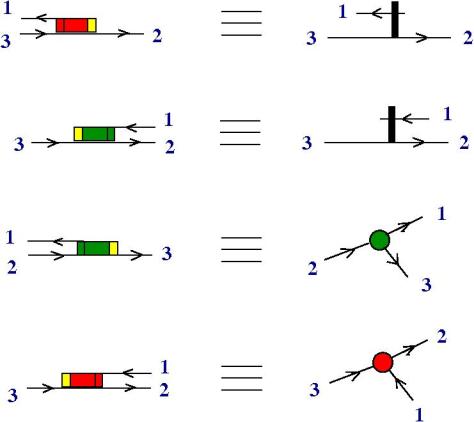
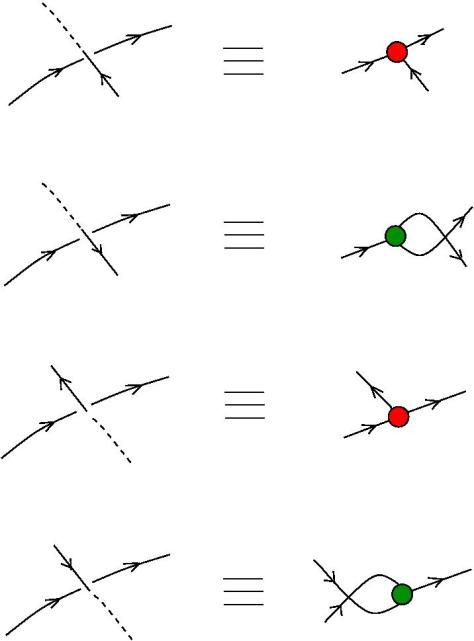
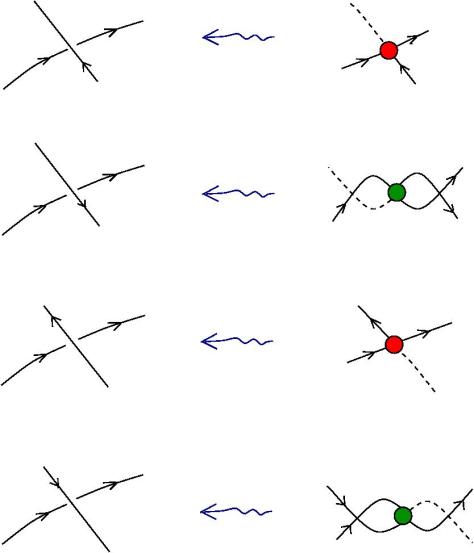
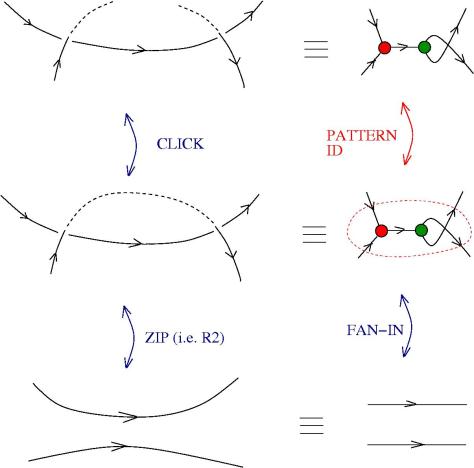
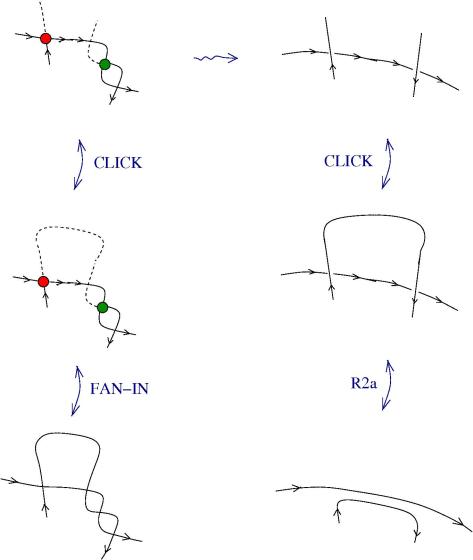
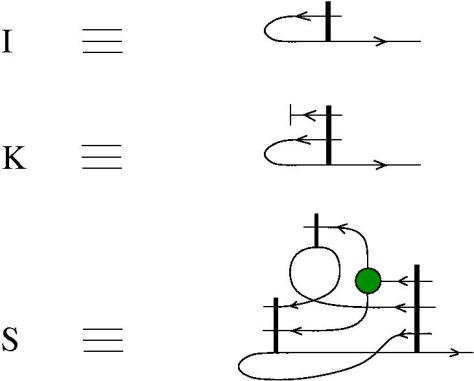
Notation discussion. We can associate a knot quandle to a knot diagram, simply by naming the arcs, then we get a presentation of a quandle. The presentation of a quandle is invariant wrt the permutation of relation or the renaming of the arcs. There is a problem, for example when an arc passes over in two crossings, we have a hidden fanout. The solution is to use a different notation and FIN (fanin) and FO (fanout) nodes. This turns the presentation into a mol file.
Can we compute with that?
Theorem: If there is a computable parser from lc to knot diagrams, such that there is a term sent to a diagram of the unknot, then all lambda terms are sent to the unknot.
We can compute with knot diagrams, but in a stupid way: if we use diagrams as a notational device. Example with the knot TM (flattened unknot, you may add the variations with the head near the tape, or the SKI calculus, to argue that it can be done in a local way. Develop this.)
Conclussion, you need a little something more, by reconnecting the arcs.
Let’s pass to ZSS
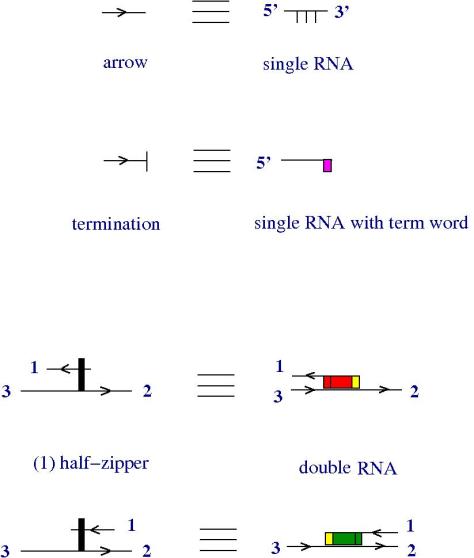
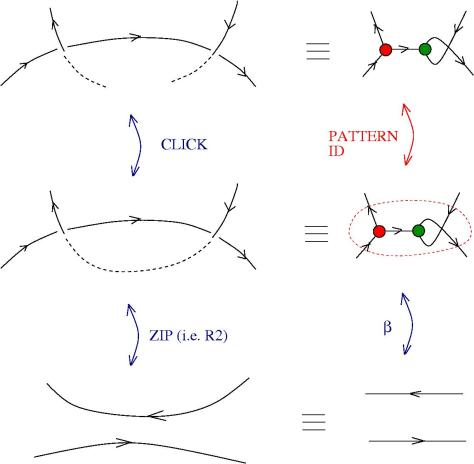
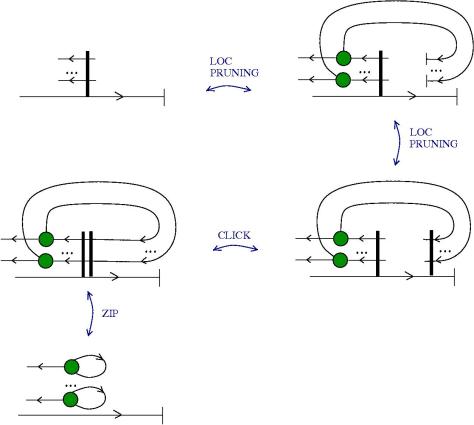
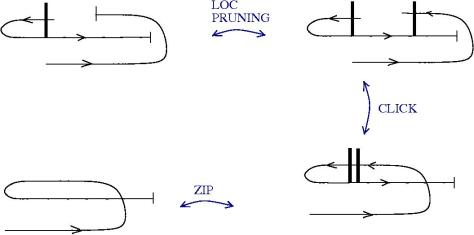
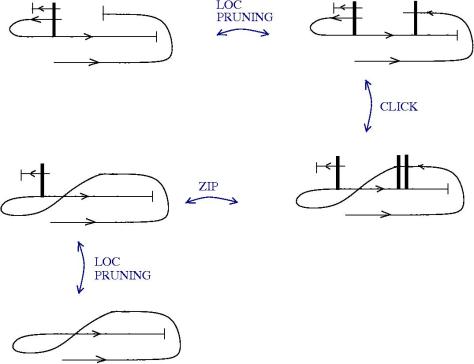
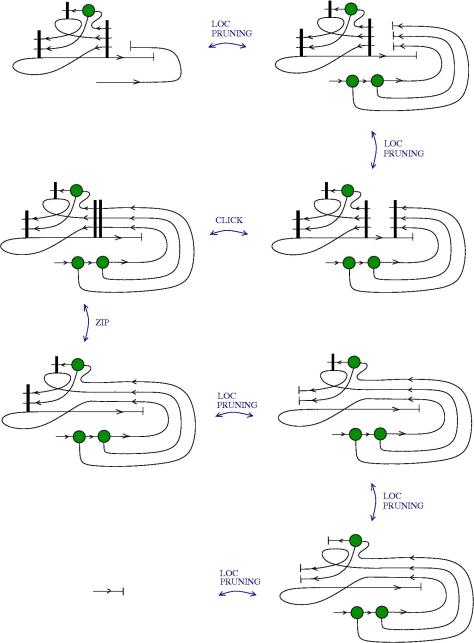
For this I introduce zippers. The idea is that a zipper is a thing which has two non-identical parts, so it’s made of two half-zippers, which are not identical.
We can use zippers like this: we have a tangle 1, a zipper, then tangle two. Explanation of the zip rewrite.
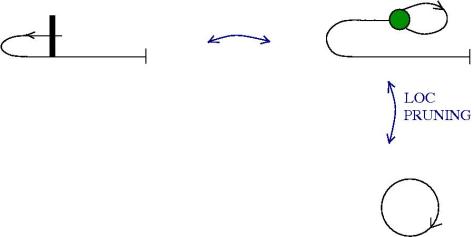
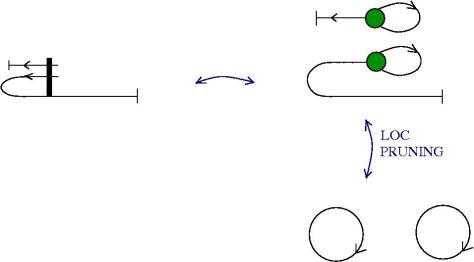
This move transforms zippers to nothing.
The move smash create zippers. Explanation of smash.
Then we have a slip move.
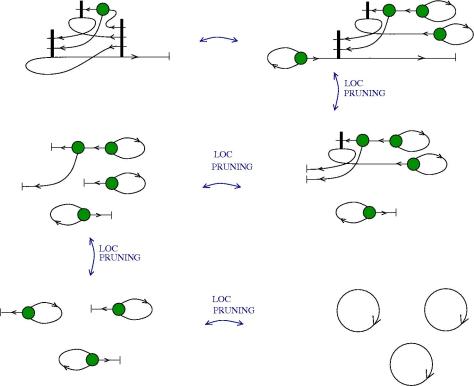
Here is the new proposal for ZL. The initial proposal used tangles with zippers, but there were also fanins and fanout nodes.
The new proposal is that we have 4 kinds of 1valent nodes, the teeths of the zippers, then we have 4 kinds of half-zippers, and then we have two kinds of crossings.
Explanation of the new proposal.
Theorem: ZSS system turns out to be able to implement dirIC, so it is universal.
Explanation of dirIC: https://mbuliga.github.io/quinegraphs/ic-vs-chem.html#icvschem










![KeepCalmStudio.com-Shortest-Explanation-Of-Chemlambda-[Knitting-Crown]-Keep-Calm-And-Use-Rna-For-Interaction-Nets](https://chorasimilarity.files.wordpress.com/2017/07/keepcalmstudio-com-shortest-explanation-of-chemlambda-knitting-crown-keep-calm-and-use-rna-for-interaction-nets.png)