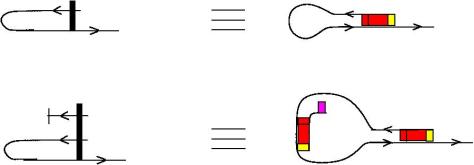UPDATE: compare with the recent Chemlambda strings (working version).
Zipper logic is a variant of chemlambda, enhanced a bit with a pattern identification move called CLICK.
Or, chemlambda is an artificial chemistry. Does it have a natural, real counterpart?
It should. Finding one is part of the other arm of the research thread presented here, which aims to unite (the computational part of) the real world with the virtual world, moved by the same artificial life/real life basic bricks. The other arm is distributed GLC.
A lot of help is needed for this, from specialists!Thank you for pointing to the relevant references, which I shall add as I receive them.
Further is a speculation, showing that zipper graphs can be turned into something which resembles very much with RNA pseudoknots.
(The fact that one can do universal computation with RNA pseudoknots is not at all surprising, maybe with the exception that one can do functional style programming with these.)
It is a matter of notation changes. Instead of using oriented crossings for denoting zippers, like in the thread called “curious crossings” — the last post is Distributivity move as a transposition (curious crossings II) — let’s use another, RNA inspired one.
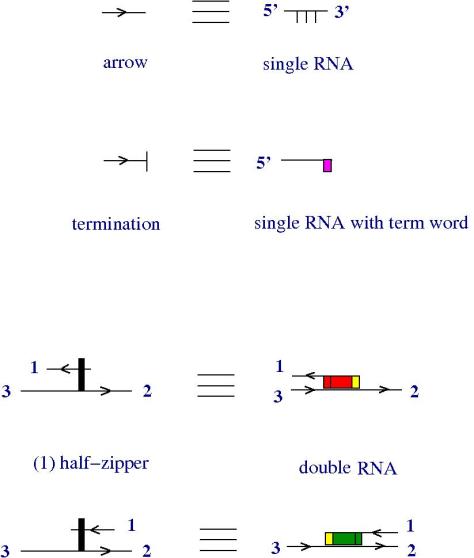
So:
- arrows (1st row) are single stranded RNA pieces. They have a natural 5′-3′ orientation
- termination node (2nd row) is a single strand RNA with a “termination word on it”, figured by a magenta rectangle
- half-zippers are double strands of RNA (3rd and 4th rows). The red rectangle is (a pair of antiparallel conjugate) word(s) and the yellow rectangle is a tag word.
Remark that any (n) half-zipper can be transformed into a sequence of (1) half-zippers, by repeated applications if the TOWER moves, therefore (n) half-zippers appear in this notation like strings of repeated words.
Another interesting thing is that on the 3rd row of the figure we have a notation for the lambda abstraction node and on the 4th row we have one of the application node form chemlambda. This invites us to transform the other two nodes — fanin and fanout — of chemlambda into double stranded RNA. It can be done like this.
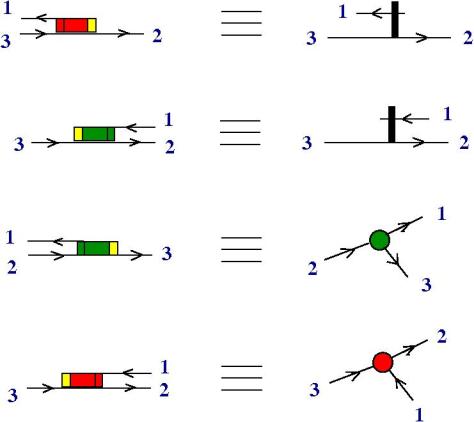
On the first two rows of this picture we see the encoding of the (1) half-zippers, i.e. the encoding of the lambda abstraction and application, as previously.
On the 3rd row is the encoding of the fanout node and on the 4th row the one of the fanin.
In this way, we arrived into the world of RNA. The moves transform as well into manipulations of RNA strings, under the action of (invisible and to be discovered) enzymes.
But why RNA pseudoknots? It is obvious, as an exercise look how the zipper I and K combinators look in this notation.
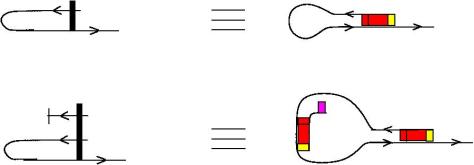
We see hairpins.
___________________________________
Conclusion: help needed for chemistry experts. Low hanging fruits here.
___________________________________