EDIT (2020): History of chemlambda (arXiv) (github) (figshare) -Graph rewrites, from graphic lambda calculus, to chemlambda, to directed interaction combinators
EDIT (2020): Chemlambda page is the place to go, in pdf form arXiv:2003.14332 [cs.AI]
EDIT (2020): See the collection of animations salvaged from G+ now available in an enhanced form.
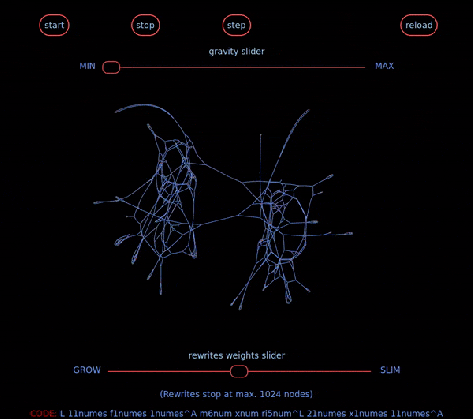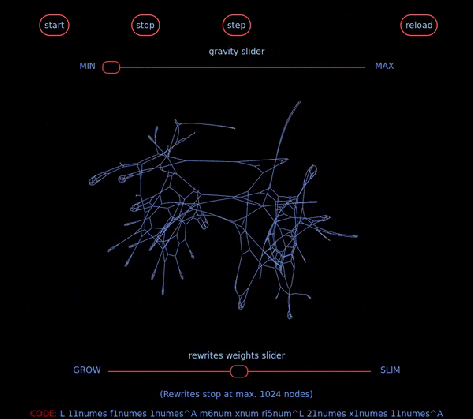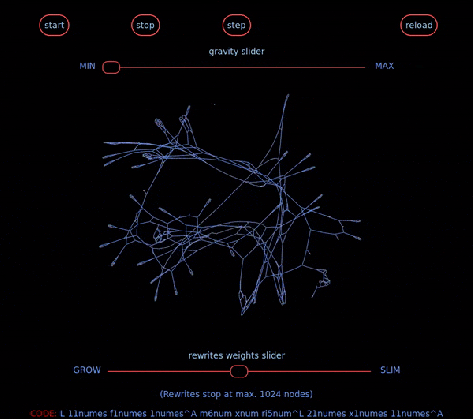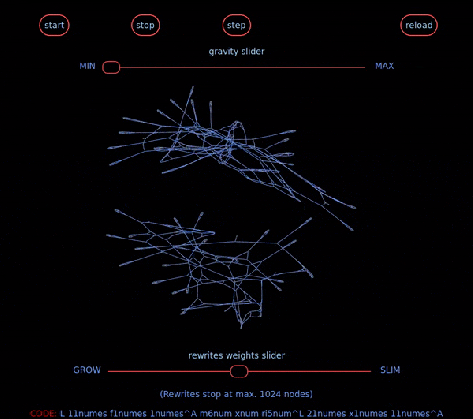
… aka Chemical Concrete Machine is a new kind of artificial chemistry.
EDIT (2019): If you want to use this blog for learning more about graphic lambda calculus and chemlambda, then you should use chemlambda.github.io.
You may read the more recent explanatory posts, starting with this one. There is also a page dedicated to chemlambda v2.
EDIT2: introduced “chemlambda strings”, eliminates enzymes, is conservative: description here.
EDIT: what you see here is chemlambda v1, for the more recent v2 see chemlambda.github.io.
____________________________________________
To read:
- (arXiv) (github) (figshare) -Graph rewrites, from graphic lambda calculus, to chemlambda, to directed interaction combinators
- chemlambda.github.io
- M. Buliga, Molecular computers, 2015. This is the article which contains the rewrites for chemlambda v2. It is also one of the first articles which “run in the browser”, besides that it offers means for validation instead of the social practice of peer review. Text version: arXiv:1811.04960
- M. Buliga, Chemical concrete machine, (DOI) arXiv:1309.6914
- M. Buliga, Graphic lambda calculus, Complex Systems 22, 4 (2013), 311-360, arXiv:1305.5786
- M. Buliga, L.H. Kauffman, GLC actors, artificial chemical connectomes, topological issues and knots, arXiv:1312.4333
- M. Buliga, L.H. Kauffman, Chemlambda, universality and self-multiplication, arXiv:1403.8046 , MIT Press article, presented by Louis in the ALIFE 14: The Fourteenth International Conference on the Synthesis and Simulation of Living Systems, July 30th – Aug 2nd 2014, New York
- Distributed GLC project.
____________________
UPDATE: See the 7 expository posts on chemlambda described with the help of g-patterns:
- part I
- part II (definition of g-patterns)
- part III (definition of moves)
- part IV (g-patterns for lambda terms 1st part)
- part V (g-patterns for lambda terms 2nd part)
- part VI (about self-multiplication of g-patterns)
- part VII (an example of reduction) .
____________________
What is the chemical concrete machine. It is a graph rewriting system, derived from graphic lambda calculus (GLC), which can be seen as a model of chemical, or why not biological, computing.
It works with very specific graphs, called “molecules”, which interact in very specific ways. This is the source of the word “concrete” in the denomination, as opposed to the Chemical abstract machine by Berry and Boudol.
I hope it can be implemented in reality, but nevertheless it can be seen as a proof of principle for the fact that the chemistry of a very small number of molecules can manifest Turing completeness. In this respect, the chemical concrete machine appears as applied Algorithmic Chemistry in a virtual world with chemistry rules which are made up by a mathematician.
For a more detailed motivation see Extreme functional programming done with biological computers
____________________
Molecules and enzymes. In the following I shall use the name “molecule” in a very loose sense. A molecule is seen as made by other smaller molecules (for example atoms are molecules) which are connected by chemical bonds, or by other molecules called “arrows”. Therefore, a molecule is seen as a graph with nodes which are smaller molecules and arrows which connect those nodes.
There might exist molecules with free arrows, or bonds.
Besides molecules, I shall also use “enzymes”. A chemical reaction will always involve two molecules, or a molecule and an enzyme. The product of the reaction is a molecule plus garbage, denoted “GARB”, which can be a collection of molecules which are inactive (i.e. they cannot be part of any other chemical reaction).
We shall work also with a list of “other molecules”, which can react (or not) one with another, this is left to the choice of the user of the chemical concrete machine.
Besides this bag of names of molecules, we have a short list of essential molecules, which are the main ingredients of the machine.
For convenience, arrows and loops (i.e. closed arrows) are seen as molecules.
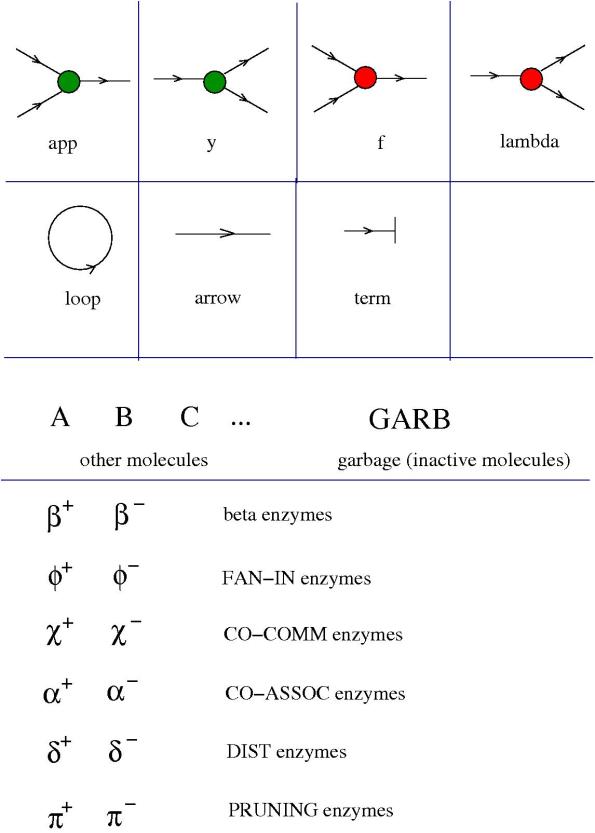
All this is described in the following figure.
On the first row you see the list of essential molecules, named respectively
- “app”, corresponding to the application gate in (graphic) lambda calculus
- “y” , corresponding to the FAN-OUT gate in graphic lambda calculus
- “lambda”, corresponding to the lambda abstraction gate in (graphic) lambda calculus
- “f” , corresponding to the
gate in graphic lambda calculus
On the second row we see a loop, an arrow (which are considered molecules, as written before), and a molecule called “term”, which corresponds in graphic lambda calculus to the termination gate.
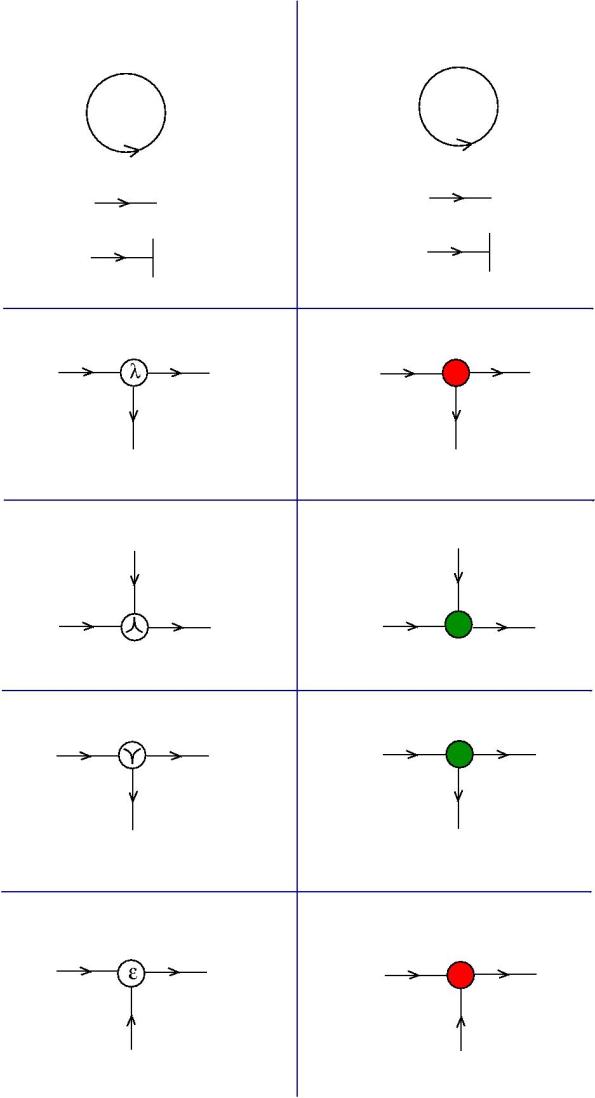
The translation from graphic lambda calculus notation to this new coloured notation is the following.
Each of these gates is a molecule.
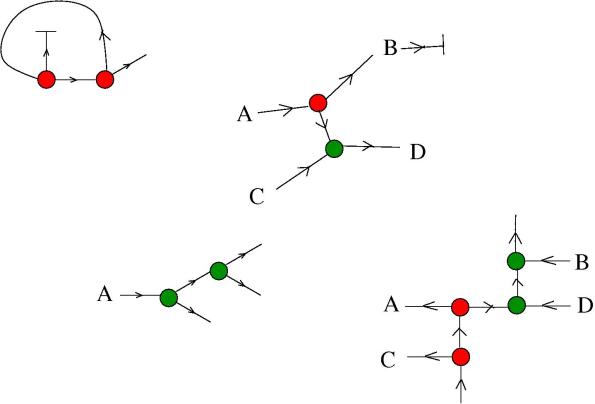
With molecules and essential molecules we build larger molecules. The physical, concrete way of constructing such molecules is left unspecified (although it can be done in the formalism of the chemical concrete machine). Here are some examples of larger molecules.
We imagine that all molecules float inside a 3D container. There may be several copies of the same molecule in the container.
Generally, the rule of building molecules is that they are made by other molecules and essential ones, simply by connecting the arrows and respecting the arrows orientations. More technically, with the correspondences specified previously, the rules of building molecules are exactly the ones for building graphs in the set described in the post Introduction to graphic lambda calculus. The only new thing is that we admit also a list
of unspecified nodes with unspecified valences, which model “other molecules”.
_____________________
So, in this simplified view of reality, molecules are such graphs. Let us see how they react with enzymes. Enzymes are not molecules, in this model. Instead, we follow the
Principle: each graphic lambda calculus gate is a molecule, each graphic lambda calculus move is an enzyme.
What is to be noticed is that any enzyme comes in two flavors: “+” and “-“. The reason for this is the following. Any chemical reaction which involves a molecule and an enzyme will correspond to a move in the list of moves of the chemical concrete machine . If you examine these moves then you shall notice that they go in both directions. Therefore, such a move appears as a pair of unidirectional moves. By convention, the moves from left to right are denoted by “+” and the moves from right to left are denoted by “-“. To each such move is associated an enzyme.
We cannot simply write reactions as
molecules + enzyme = molecules + GARB
because these molecules may be complicated beasts (graphs) and because, as we shall see, the enzymes prefer to react with certain patterns (subgraphs) of molecules. For specifying how the reaction takes place we need:
- molecules
- an enzyme
- and a “reaction site”, which is a small part of the initial collection of molecules.
The reaction site have to be present in the molecule, otherwise the reaction cannot happen.
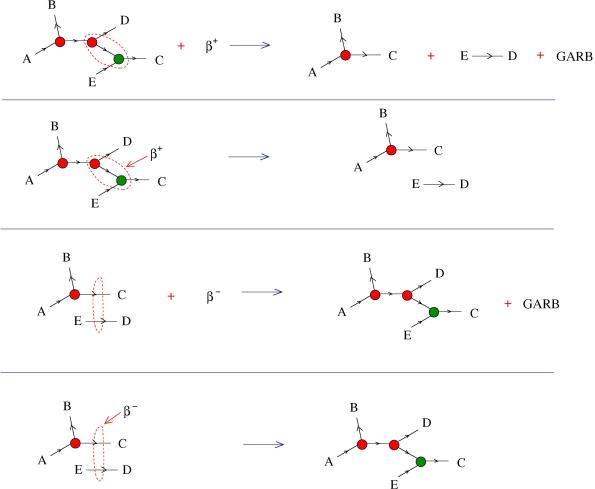
In the following figure I consider two examples of reactions (which correspond to graphic beta moves in the realm of graphic lambda calculus).
In the first row we see a reaction between a molecule and the enzyme , which results into two other molecules and some GARB. There is a small region in the initial molecule, marked by a dashed red closed curve, which represents a reaction site for the
molecule.
In the second row is written the same reaction, but in a simpler form. The red “+” sign is eliminated, the two molecules which are obtained are juxtaposes, as if they are floating in the 3D container, and the GARB is ignored. Moreover, the enzyme points towards the reaction site.
The rows 3 and 4 describe another reaction. At closer inspection, it’s a reaction which can be interpreted as the inverse of the first one. Let’s examine directly the 4th row (which is obtained from the 3rd row by the same procedure as the 2nd row was obtained from the 1st). The reaction site of the enzyme is a pair of arrows from two different molecules, The resulting molecule is the same as the initial molecule from the previous reaction.
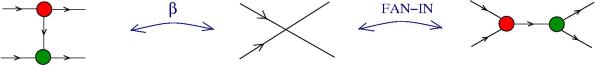
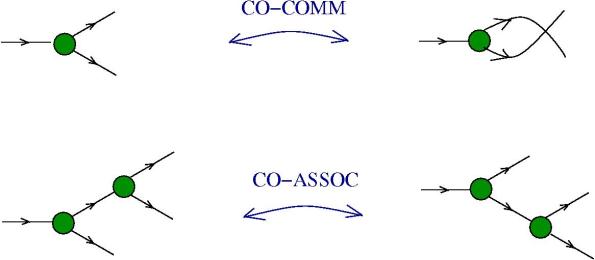
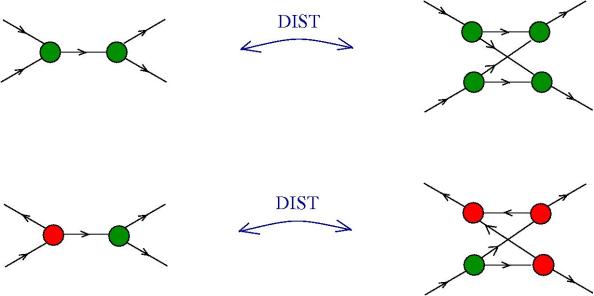
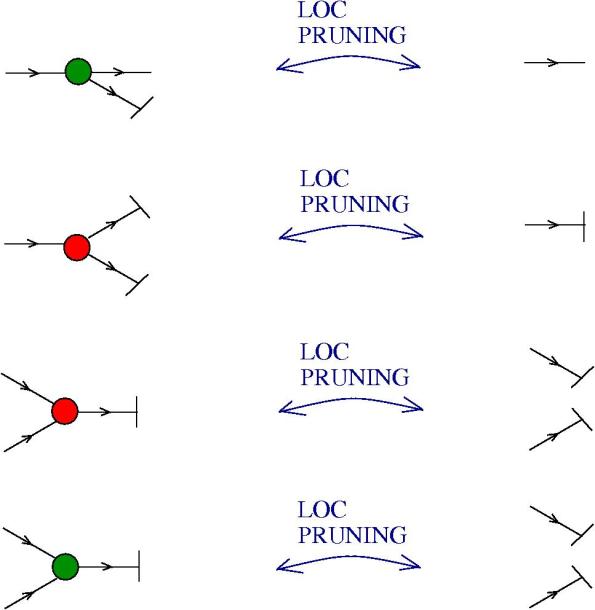
The moves. Each move is made possible by an enzyme (hence move = enzyme). Here is the list, with the names taken from graphic lambda calculus, by using the dictionary for translation. The DIST and FAN-IN moves are not part of the graphic lambda calculus. Their addition eliminates the need for global moves, therefore the chemical concrete machine has only local moves! Moreover the FAN-IN move replaces the emergent algebra moves from graphic lambda calculus, therefore giving to the epsilon gate another behaviour.
- graphic beta move and FAN-IN:
- local FAN-OUT moves:
- DIST moves:
- LOC PRUNING moves:
- Elimination of loops:
_______________
Let’s see what the chemical concrete machine can do.
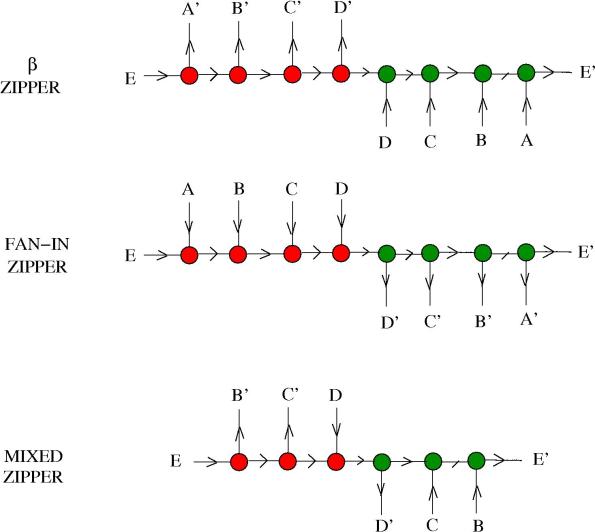
1. Lists and locks. Suppose you have a family of molecules which you want to free them in the medium in a given order. This corresponds to having a list of molecules, which is “read” sequentially. I shall model this with the help of the zipper from graphic lambda calculus.
Suppose that the molecules we want to manipulate have the form , with
and
from the family of “other molecules” and
an arrow, in the model described in Chemical concrete machine, detailed (I). Here are three zippers (lists).
The first zipper, called a zipper, behaves in the following way. In the presence of
enzymes, there is only one reaction site available, namely the one involving the red and green nodes in the neighbourhood of the
. So there is only one reaction possible with a
enzyme, which has a a result the molecule
and a new, shorter
zipper. This new zipper has only one reaction site, this time involving nodes in the neighbourhood of
, so the reaction with the enzyme
gives
and a new, shorter zipper. The reaction continues like this, freeing in order the molecules
, then
and
.
The second zipper is called a FAN-IN zipper (or a zipper). It behaves the same as the previous one, but this time in the presence of the FAN-IN enzyme
.
On the third row we see a mixed zipper. The first molecule is released only in the presence of a
enzyme, then we are left with a
zipper.
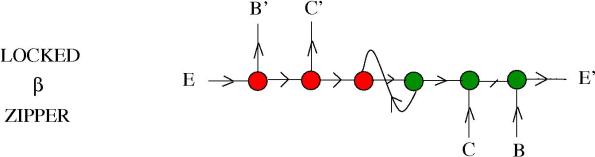
This can be used to lock zippers. Look for example at the following molecule:
called a locked zipper. In the presence of only
enzymes, nothing happens. If we add into the reactor also
enzymes, then the zipper unlocks, by releasing a loop (that’s seen as garbage) and a
zipper which starts to react with
enzymes.
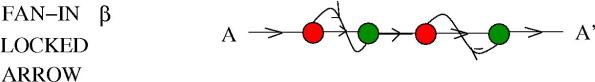
The same idea can be used for keeping a molecule inactive unless both and
enzymes are present in the reactor. Say that w have a molecule
which is made inactive under the form presented in the following figure
The molecule is locked, but it has two reaction sites, one sensible to , the other sensible to
. Both enzymes are needed for unlocking the molecule, but there is no preferred order of reaction with the enzymes (in particular these reactions can happen in parallel).
_____________________
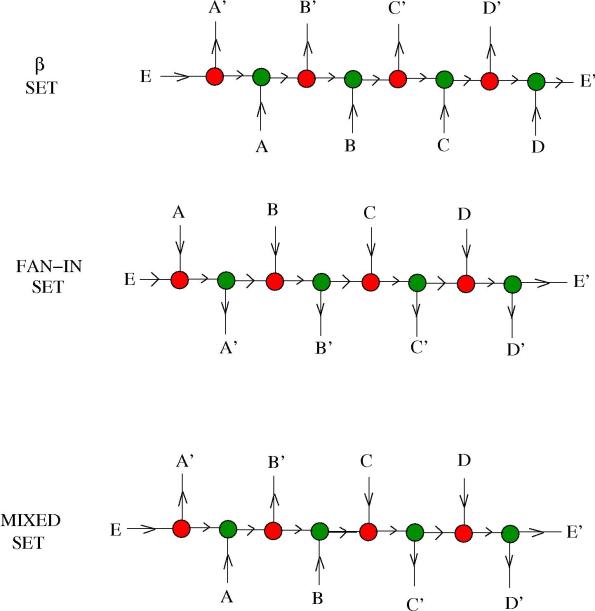
2. Sets. Suppose now that we don’t want to release the molecules in a given order. We need to prepare a molecule which has several reaction sites available, so that multiple reactions can happen in parallel, as in the last example. Mathematically, that could be seen as a representation of the set of molecules we want to free, instead of the list of them. This is easy, as described in the next figure:
On the first row we see what is called a set. It has 4 possible reaction sites with the enzyme
, therefore, in the presence of this enzyme, the molecules
, … , $E \rightarrow E’$ are released at the same moment. Alternatively, we may think about a
set as a bag of molecules which releases (according to the probability of the reaction with a
enzyme$) one of the four molecules
, … ,
, at random. (It should be interesting to study the evolution of this reaction, because now there are only 3 reaction sites left, …)
On the second row we see a FAN-IN, or set. It behaves the same as the previous one, but this time in the presence of the FAN-IN
enzyme.
Finally, we see a mixed set on the third row. (It should have an interesting dynamics, as a function of the concentrations of the two enzymes.)
_____________________
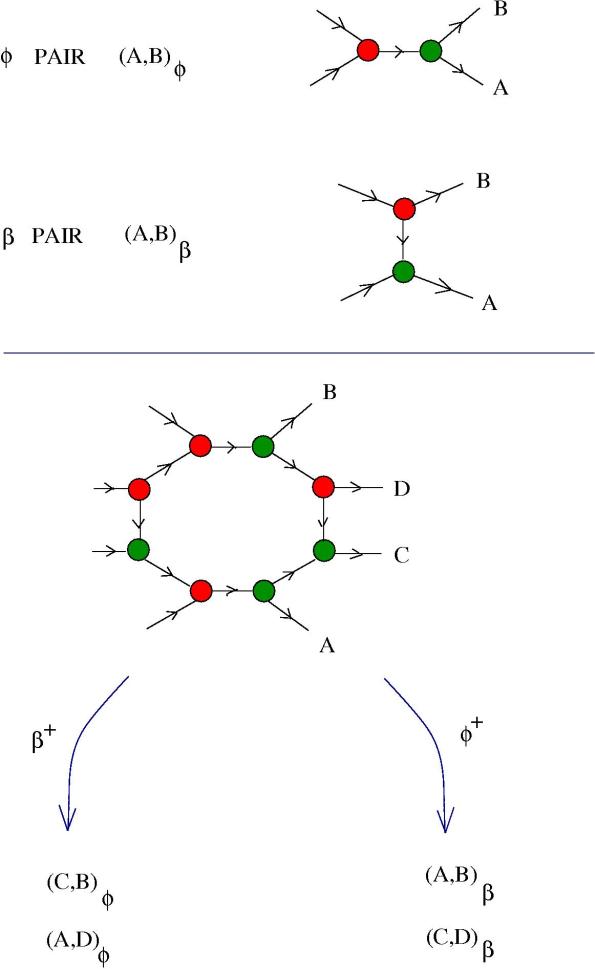
3. Pairs. Actually, there is no limit but the imagination to what geometrical operations to consider, see for example the posts Sets, lists and order of moves in graphic lambda calculus and Pair of synapses, one controlling the other (B-type NN part III) . As another example, here is a more involved molecule, which produces different pairs of molecules, according to the presence of or
enzymes.
In the following figure we see how we model a pair of molecules, then two possible reactions a re presented.
The idea is that we can decide, by controlling the amount of or
, to couple
with
and
with
, or to couple
with
and
with
. Why? Suppose that
and
can react with both
and
, depending of course on how close available molecules are.
For example, we want that and
molecules to be, statistically, one in the proximity of another. We add to the reactor some enzyme
and we obtain lots of pairs
. Now, when we add further the
enzyme, then, immediately after the reaction with this enzyme we are going to have lots of pairs of molecules
and
which are physically close one to another, starting to react.
Instead, if we first introduce enzymes and then
enzymes, then
would react more likely with
.
_____________________
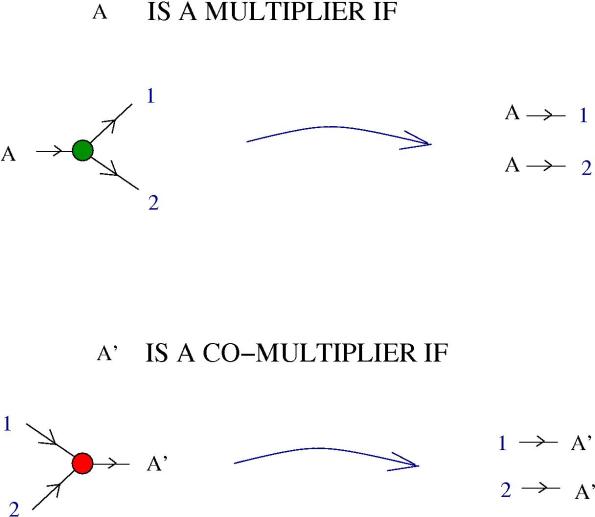
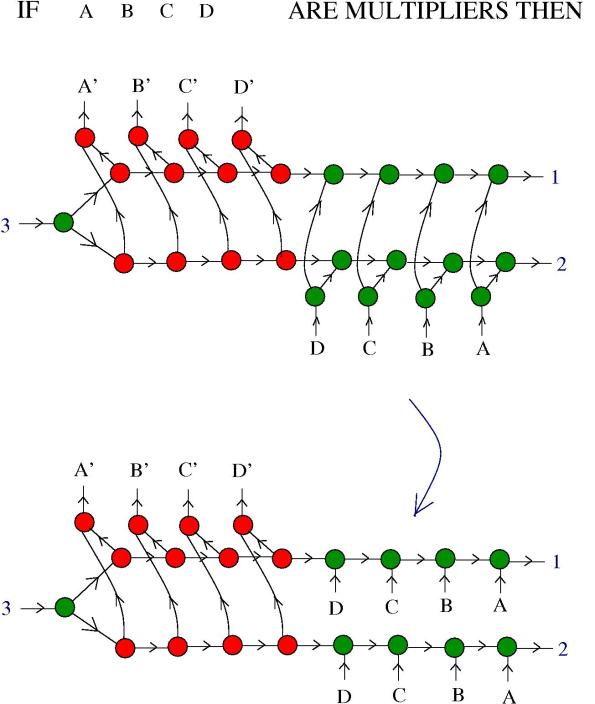
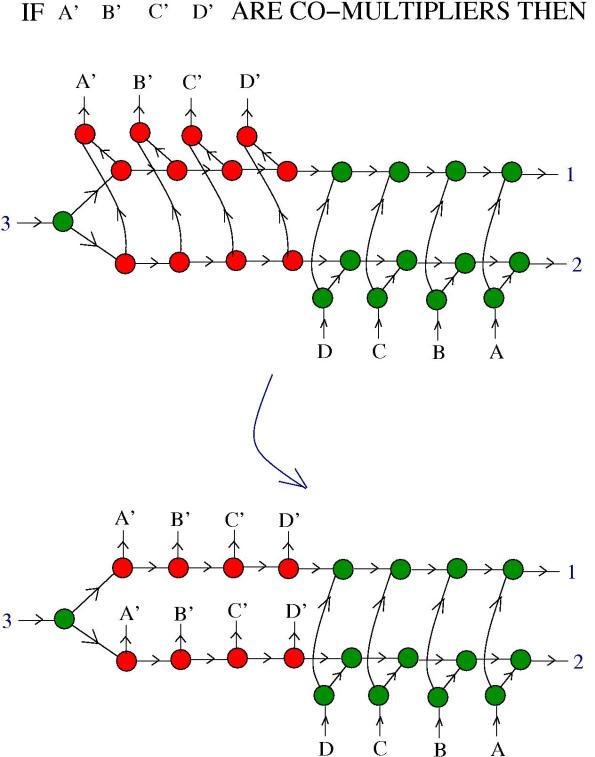
4. Multipliers and comultipliers. Basically, multipliers and co-multipliers are molecules which self-multiply. More precisely, in the next figure we see the definition of those:
Here and
are molecules from the formalism of the chemical concrete machine and
and
are labels. The blue arrow means any chemical reaction which is allowed.
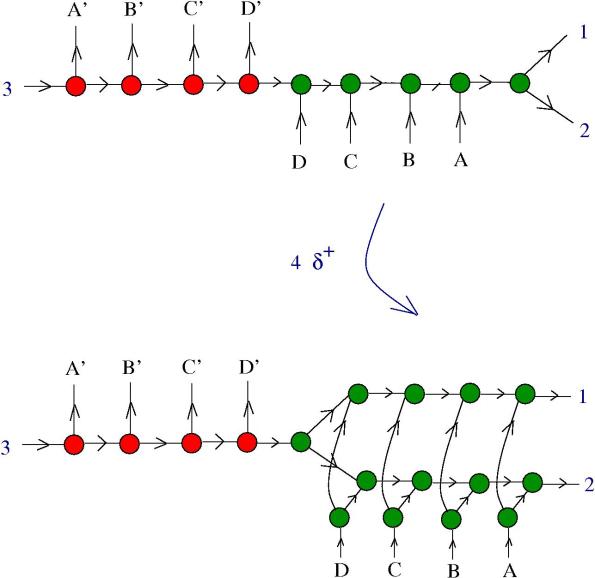
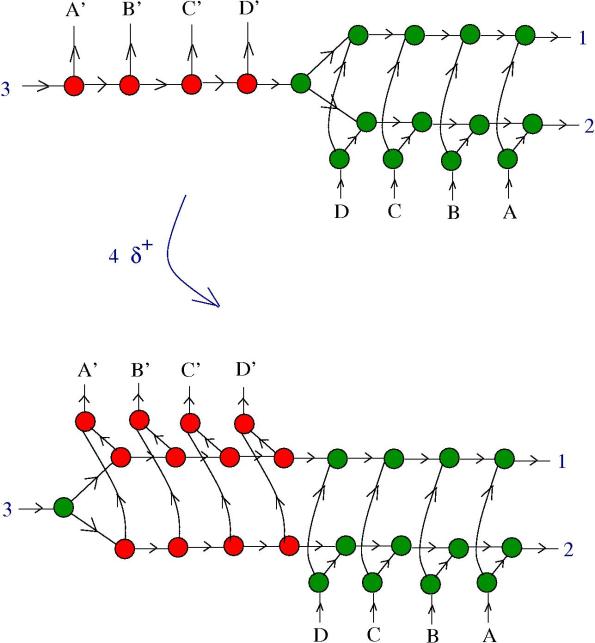
5. Zippers and multipliers. We want to know if a zipper can be a multiplier. In the following figure we see what happens in the presence of DIST enzymes:
The reaction continues:
Now, the zipper multiplied into two zippers, but they are still connected. We need more information about and
. Remark that:
In conclusion: if are multipliers and
are co-multipliers, then the zipper is a multiplier!
6. Turing universality. Read the following posts:

… the way our DNA would prove it …