The replication mechanism for circular bacterial chromosomes is known. There are two replication forks which propagate in two directions, until they meet again somewhere and the replication is finished.
[source, found starting from the wiki page on circular bacterial chromosomes]
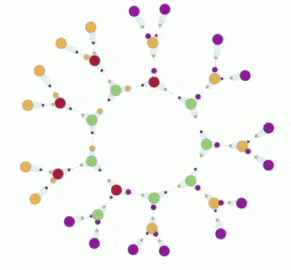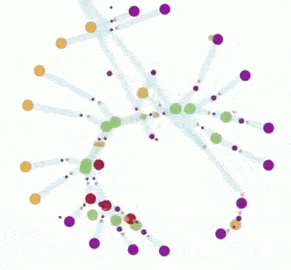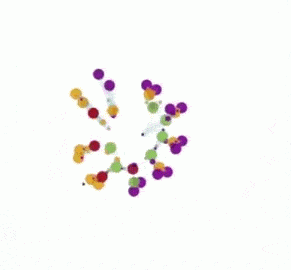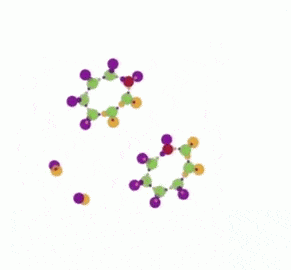
In the artificial chemistry chemlambda something similar can be done. This leads to some interesting questions. But first, here is a short animation which describes the chemlambda simulation.
The animation has two parts, where the same simulation is shown. In the first part some nodes are fixed, in order to ease the observation of the propagation of the replication, which is like the propagation of the replication forks. In the second part there is no node fixed, which makes easy to notice that eventually we get two ring molecules from one.
____________
If the visual evidence convinced you, then it is time to go to the explanations and questions.
But notice that:
- The replication of circular DNA molecules is done with real chemistry
- The replication of the circular molecule from the animation is done with an artificial chemistry model.
The natural question to ask is: are these chemistries the same?
The answer may be more subtle than a simple yes or no. As more visual food for thinking, take a look at a picture from the Nature Nanotechnology Letter “Self-replication of DNA rings” http://www.nature.com/nnano/journal/vaop/ncurrent/full/nnano.2015.87.html by Junghoon Kim, Junwye Lee, Shogo Hamada, Satoshi Murata & Sung Ha Park
[this is Figure 1 from the article]
This is a real ring molecule, made of patterns which, themselves are made of DNA. The visual similarity with the start of the chemlambda simulation is striking.
But this DNA ring is not a DNA ring as in the first figure. It is made by humans, with real chemistry.
Therefore the boldfaced question can be rephrased as:
Are there real chemical assemblies which react as of they are nodes and bonds of the artificial chemistry?
Like actors in a play, there could be a real chemical assembly which plays the role of a red atom in the artificial chemistry, another real chemical assembly which plays the role of a green atom, another for a small atom (called “port”) and another for a bond between these artificial chemistry atoms.
From one side, this is not surprising, for example a DNA molecule is figured as a string of letters A, C, T, G, but each letter is a real molecule. Take A (adenine)
[source]
Likewise, each atom from the artificial chemistry (like A (application), L (lambda abstraction), etc) could be embodied in real chemistry by a real molecule. (I am not suggesting that the DNA bases are to be put into correspondence with artificial chemistry atoms.)
Similarly, there are real molecule which could play the role of bonds. As an ilustration (only), I’ll take the D-deoxyribose, which is involved into the backbone structure of a DNA molecule.
[source]
So it turns out that it is not so easy to answer to the question, although for a chemist may be much easier than for a mathematician.
___________
0. (few words about validation) If you have the alternative to validate what you read, then it is preferable to authority statements or hearsay from editors. Most often they use anonymous peer-reviews which are unavailable to the readers.
Validation means that the reader can make an opinion about a piece of research by using the means which the author provides. Of course that if the means are not enough for the reader, then it is the author who takes the blame.
The artificial chemistry animation has been done by screen recording of the result of a computation. As the algorithm is random, you can produce another result by following the instructions from
https://github.com/chorasimilarity/chemlambda-gui/blob/gh-pages/dynamic/README.md
I used the mol file model1.mol and quiner.sh. The mol file contains the initial molecule, in the mol format. The script quiner.sh calls the program quiner.awk, which produces a html and javascript file (called model1.html), which you can see in a browser.
I added text to such a result and made a demo some time ago
http://chorasimilarity.github.io/chemlambda-gui/dynamic/model1.html
(when? look at the history in the github repo, for example: https://github.com/chorasimilarity/chemlambda-gui/commits/gh-pages/dynamic/model1.html)
1. Chemlambda is a model of computation based on artificial chemistry which is claimed to be very closed to the way Nature computes (chemically), especially when it comes to the chemical basis of computation in living organisms.
This is a claim which can be validated through examples. This is one of the examples. There are many other more; the chemlambda collection shows some of them in a way which is hopefully easy to read (and validate).
A stronger claim, made in the article Molecular computers (link further), is that chemlambda is real chemistry in disguise, meaning that there exist real chemicals which react according to the chemlambda rewrites and also according to the reduction algorithm (which does random rewrites, as if produced by random encounters of the parts of the molecule with invisible enzymes, one enzyme type per rewrite type).
This claim can be validated by chemists.
http://chorasimilarity.github.io/chemlambda-gui/dynamic/molecular.html
2. In the animation the duplication of the ring molecule is achieved, mostly, by the propagation of chemlambda DIST rewrites. A rewrite from the family DIST typically doubles the number of nodes involved (from 2 to 4 nodes), which vaguely suggest that DIST rewrites may be achieved by a DNA replication mechanism.
(List of rewrites here:
http://chorasimilarity.github.io/chemlambda-gui/dynamic/moves.html )
So, from my point of view, the question I have is: are there DNA assemblies for the nodes, ports and bonds of chemlambda molecules?
3. In the first part of the animation you see the ring molecule with some (free in FRIN and free out FROUT) nodes fixed. Actually you may attach more graphs at the free nodes (yellow and magenta 1-valent nodes in the animation).
You can clearly see the propagation of the DIST rewrites. In the process, if you look closer, there are two nodes which disappear. Indeed, the initial ring has 9 nodes, the two copies have 7 nodes each. That is because the site where the replication is initiated (made of two nodes) is not replicated itself. You can see the traces of it as a pair of two bonds which connect, each, a free in with a free out node.
In the second part of the animation, the same run is repeated, this time without fixing the FRIN and FROUT nodes before the animation starts. Now you can see the two copies, each with 7 nodes, and the remnants of the initiation site, as a two pairs of FRIN-FROUT nodes.
_________________________________________